Taconic Biosciences' Genetics Sciences team perform state-of-the-art molecular characterization assays throughout the breeding of every colony to ensure that animal model integrity and quality are maintained.
Reliable results depend on having the right tools and research materials. Taconic helps ensure your mouse or rat models have the desired genetic profile through a variety of assays and techniques.
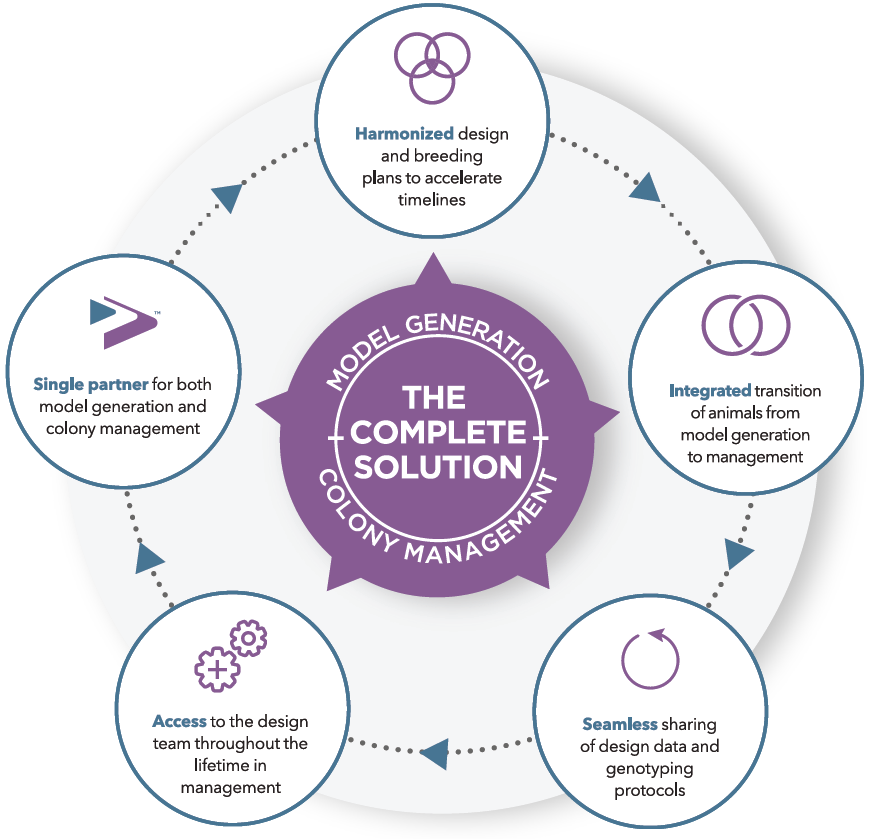
During your consultation with the Project team, we will create an appropriate genotyping and molecular analysis strategy to support your study.










.jpg)

.jpg)
.jpg)
.jpg)
.jpg)



.jpg)


.jpg)
.jpg)


.jpg)



.jpg)

.jpg)