 In a recent review article, David Masopust and his coauthors from the University of Minnesota Medical School discuss the relevancy of mouse models for use in immunological studies and translatability of immune system studies using SPF mice. Of Mice, Dirty Mice, and Men: Using Mice to Understand Human Immunology1 provides a short introduction to the development of the laboratory mouse while calling out a number of significant scientific advances that came out of research utilizing laboratory mice, including the discovery of IgG in the 1930s, understanding acquired immunologic tolerance in the 1950s, and, most recently, the discovery of checkpoint blockade therapy.
In a recent review article, David Masopust and his coauthors from the University of Minnesota Medical School discuss the relevancy of mouse models for use in immunological studies and translatability of immune system studies using SPF mice. Of Mice, Dirty Mice, and Men: Using Mice to Understand Human Immunology1 provides a short introduction to the development of the laboratory mouse while calling out a number of significant scientific advances that came out of research utilizing laboratory mice, including the discovery of IgG in the 1930s, understanding acquired immunologic tolerance in the 1950s, and, most recently, the discovery of checkpoint blockade therapy.
From SPF to Dirty Mice
Specific Pathogen Free (SPF) mice date back to the late 1950s, when the term was first used to describe the microbiological status (i.e., health) of mouse colonies in terms of the presence or absence of a specific list of pathogens2,3. While there is recognition of what SPF means in a broad sense, the list of defined organisms varies from lab to lab. "Evidence that microbial experience impacts the immune response of laboratory mice is compelling," Masopust and his coauthors conclude. "At its most extreme, the development of gnotobiotic, or germ-free, mice has revealed the profound impact of commensal organisms on everything from host metabolism to pathogen vulnerability." Scientists have "noted that a key phenotype of a certain mouse strain is no longer reproducible upon changing laboratory locations, or simply by housing their mice in a different room at the same institution."The authors cite a well-known example with the Non Obese Diabetic (NOD) strain, in which the diabetic phenotype changed depending on the presence or absence of certain commensal organisms4,5,6.
The accumulation of evidence leads Masopust to a compelling question: "What are we missing by conducting most mouse experiments under SPF conditions? Are there practicable methods to return laboratory mice to a more physiologic level of microbial experience? If so, would such an endeavor have value?"
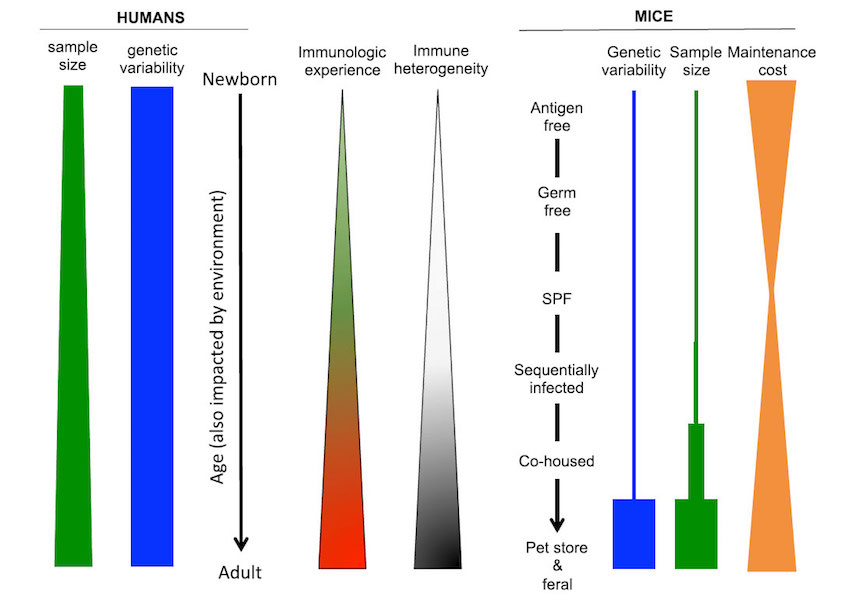
Practical considerations for research in humans and in mice with varying degrees of immune experience. Humans exhibit significant genetic variability, unlike studies that employ inbred strains of mice1.
Are Dirty Mice More Translatable?
At this point, the discussion turns to the use of "dirty mice" — feral mice, pet store mice, or mice prepared by the deliberate transfer of defined pathogens. "Data suggest[ed] that cohoused inbred mice, by having a more physiological exposure to natural mouse pathogens, were a more accurate reflection of the adult human immune system, and hence would be more relevant to translation of mouse immunology studies7,8".The takeaway message here is that microbiome matters. SPF mice have their utility in a wide spectrum of studies. However, when considering immunological studies, investigators must take time to understand how the presence or absence of specific organisms may affect study results.
In their conclusions, the authors point out the utility of laboratory mice in understanding the basics of the immune system, both past and future. They end by noting that "normalizing the immunological experience of laboratory mice [...] should be viewed as an element in the larger progression to improve the value of mice as a model for the human immune system."
 Download the Taconic Biosciences' White Papers:
Download the Taconic Biosciences' White Papers:- Practical Considerations for Fecal Transplantation Experiments in Rodents
- The Critical Role of Rodent Model Health Standards










.jpg)

.jpg)
.jpg)
.jpg)
.jpg)



.jpg)


.jpg)
.jpg)


.jpg)


.jpg)

.jpg)



