Application Areas:
Nod2
- Description
- Price & Licensing
- Overview
- Genetics
- Guides & Publications
- Transit, Housing & Welfare
- Diet
Overview
Nomenclature: C57BL/6NTac-Nod2em2Tac
- Carries a deletion of exon 3 and 4 of Nod2 gene
- The deletion removes part of the NACHT domain, resulting in a frame-shift leading to a premature stop codon in exon 5. Additionally, the resulting transcript may be a target for nonsense-mediated RNA decay and may therefore not be expressed at significant level.
- Homozygous mice should not produce any Nod2 protein.
- Useful in inflammatory bowel disease, microbiome, Parkinson's disease, immunology and inflammation research.
- Nod2 is involved in gastrointestinal immunity, it is associated with inflammatory bowel disease (IBD) (Scientific Reports 2015, 5 (1), 12018, Science 2005, 307 (5710), 734-738). Mutations in Nod2 are also found in Blau and Yao syndrome patients.
- Nod2 knockout mice have altered microbial community in the colonic mucosa when compared to Nod1 KO and wild type mice (Gut 2011, 60 (10), 1354-1362). Gut-associated lymphoid tissue (GALT) is also altered in Nod2 knockout mice resulting in increased epithelial permeability.
- A host carrying mutant NOD2 alleles may have a diminished epithelial defense against enteric bacteria (Journal of Crohns and Colitis 2016, 10 (12), 1428-1436, Nature Medicine 2016, 22 (5), 524-530).
- Loss of Nod2 gene also enhances epithelial dysplasia following chemically induced injury (Journal of Clinical Investigation 2013, 123 (2), 700-711).
- Nod2 deficiency can protect neurons against 6-hydroxydopamine (6-OHDA) induced cell death, which mimics Parkinson's disease pathology (Journal of Neuroinflammation 2018, 15 (1), 243).
Orders by weight: Taconic cannot accept orders by weight for this model. Please note that shipments may contain animals with a larger weight variation.
Origin
This model is cryopreserved and available for recovery. Models can typically be recovered and delivered to customers within 12 weeks after order receipt. Purchase of this model includes perpetual use rights and a deliverable of four mutant animals at the Murine Pathogen Free™ health standard along with a genotyping protocol. For models which include a recombinase gene or multiple alleles, all alleles will be provided, but individual animals may not contain all mutant alleles.
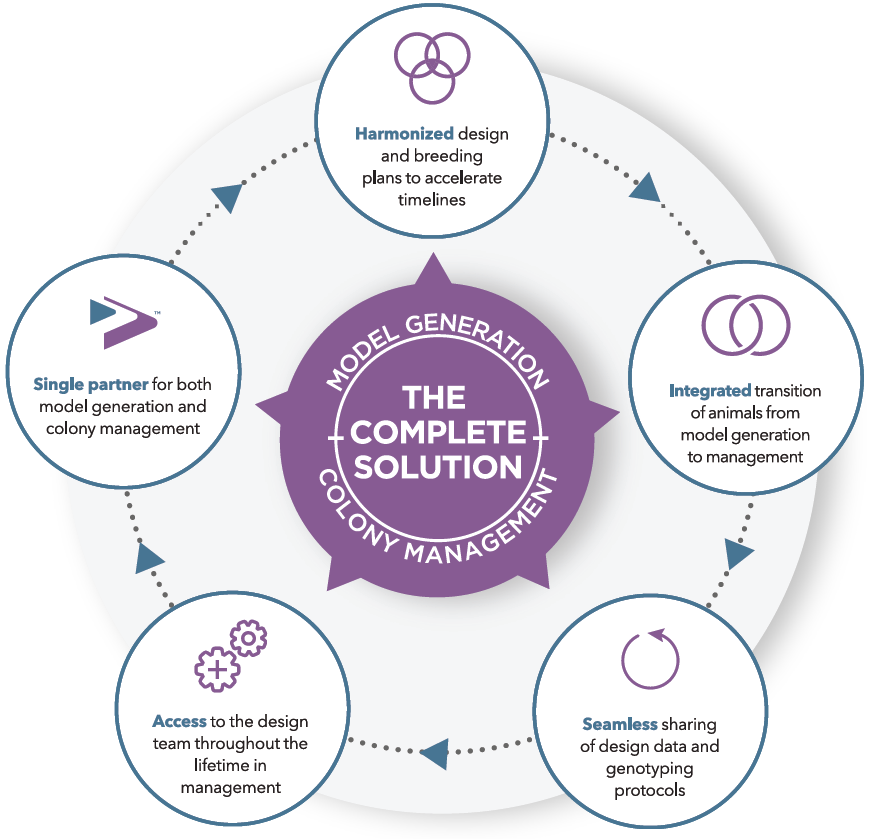
Taconic’s Colony Management experts can design a plan to grow your colony faster.
Genetics
Guides & Publications
Initial Publication: There is no specific publication describing the generation of these mice, but multiple publications exist demonstrating applications using similar models. See reference list.
Other Publications:
- Kim, H.; Zhao, Q.; Zheng, H.; Li, X.; Zhang, T.; Ma, X. Scientific Reports 2015, 5 (1), 12018.
- Maeda, S.; Hsu, L.C.; Liu, H.; Bankston, L.A.; Iimura, M.; Kagnoff, M.F.; Eckmann, L.; Karin, M. Science 2005, 307 (5710), 734-738.
- Rehman, A.; Sina, C.; Gavrilova, O.; Hasler, R.; Ott, S.; Baines, J. F.; Schreiber, S.; Rosenstiel, P. Gut 2011, 60 (10), 1354–1362.
- Nabhani, Z. A.; Lepage, P.; Mauny, P.; Montcuquet, N.; Roy, M.; Roux, K. L.; Dussaillant, M.; Berrebi, D.; Hugot, J.-P.; Barreau, F. Journal of Crohns and Colitis 2016, 10 (12), 1428–1436.
- Couturier-Maillard, A.; Secher, T.; Rehman, A.; Normand, S.; Arcangelis, A. D.; Haesler, R.; Huot, L.; Grandjean, T.; Bressenot, A.; Delanoye-Crespin, A.; Gaillot, O.; Schreiber, S.; Lemoine, Y.; Ryffel, B.; Hot, D.; Nùñez, G.; Chen, G.; Rosenstiel, P.; Chamaillard, M. Journal of Clinical Investigation 2013, 123 (2), 700-711.
- Kim, D.; Kim, Y.-G.; Seo, S.-U.; Kim, D.-J.; Kamada, N.; Prescott, D.; Chamaillard, M.; Philpott, D. J.; Rosenstiel, P.; Inohara, N.; Núñez, G. Nature Medicine 2016, 22 (5), 524–530.
- Cheng, L.; Chen, L.; Wei, X.; Wang, Y.; Ren, Z.; Zeng, S.; Zhang, X.; Wen, H.; Gao, C.; Liu, H. Journal of Neuroinflammation 2018, 15 (1), 243.
Transit, Housing & Welfare
Need more info? Click the live chat button or Contact Us
Diet
- Licensing
- Pricing - USD
- Pricing - EUR
- Pricing - DKK
- Select my Health Standard
- Get Custom Pricing Guide
Nod2
This model is sold under terms which grant perpetual use rights.
Pricing - USD
16476-EZcohort-4
| Item | Commercial | Nonprofit |
|---|---|---|
| Cryopreserved Model | US$14,000.00 | US$14,000.00 |
Cryopreserved models are invoiced upon shipment of recovered animals. Once orders are placed, the full purchase price will be applied if the order is canceled. For orders greater than 4 animals, please contact Taconic for options.
Fees for Taconic Transit Cages™ and freight are in addition to the price above.
Pricing - EUR
16476-EZcohort-4
| Item | Commercial | Nonprofit |
|---|---|---|
| Cryopreserved Model | 12.100,00 € | 12.100,00 € |
Cryopreserved models are invoiced upon shipment of recovered animals. Once orders are placed, the full purchase price will be applied if the order is canceled. For orders greater than 4 animals, please contact Taconic for options.
Fees for Taconic Transit Cages™ and freight are in addition to the price above.
Pricing - DKK
16476-EZcohort-4
| Item | Commercial | Nonprofit |
|---|---|---|
| Cryopreserved Model | kr.90.266,00 | kr.90.266,00 |
Cryopreserved models are invoiced upon shipment of recovered animals. Once orders are placed, the full purchase price will be applied if the order is canceled. For orders greater than 4 animals, please contact Taconic for options.
Fees for Taconic Transit Cages™ and freight are in addition to the price above.
Select my Health Standard
Need help choosing the right Taconic Biosciences health standard for your research?
Use the Health Standard Selector to enter your exclusion list. The tool will tell you which health standards meet your requirements.
Get custom pricing guide
Schedule A Scientific Consultation
Connect directly with a member of our Scientific Solutions team who can help you select the most appropriate model and maximize your experimental success.










.jpg)

.jpg)
.jpg)
.jpg)
.jpg)



.jpg)


.jpg)
.jpg)


.jpg)



.jpg)

.jpg)